Build a containerized PDF to Markdown converter using Docling and Streamlit ☁️

Introduction
In one of my last discussions, I was asked to help a business partner to start using Docling for document preparation to be sent to a RAG. Putting aside the project details, the application is meant to run on a Kubernetes cluster.
The sample provided here provides the steps to create a Python application using Docling document conversion capabilities, and using Streamlit to build the web UI. The application can be containerized to be deployed on a k8s cluster.
What is Docling
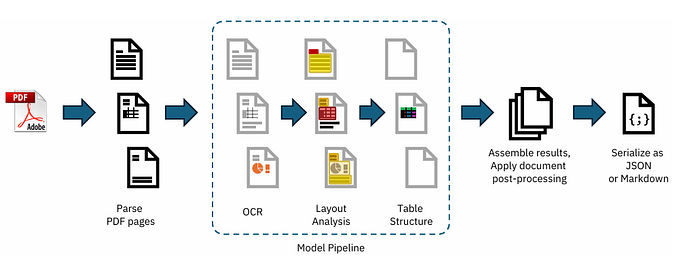
Docling simplifies document processing, parsing diverse formats — including advanced PDF understanding — and providing seamless integrations with the gen AI ecosystem.
Features
- 🗂️ Parsing of multiple document formats incl. PDF, DOCX, XLSX, HTML, images, and more
- 📑 Advanced PDF understanding incl. page layout, reading order, table structure, code, formulas, image classification, and more
- 🧬 Unified, expressive DoclingDocument representation format
- ↪️ Various export formats and options, including Markdown, HTML, and lossless JSON
- 🔒 Local execution capabilities for sensitive data and air-gapped environments
- 🤖 Plug-and-play integrations incl. LangChain, LlamaIndex, Crew AI & Haystack for agentic AI
- 🔍 Extensive OCR support for scanned PDFs and images
- 💻 Simple and convenient CLI
The sample application

Disclaimer: this is a very simple implementation, in order to showcase the capacities. It should be enhanced for a scaled / industrialized application.
On Intel machines, most likely there is a need to troubleshoot the configuration in order to make it runnable. The Conda activation will probably be required. Hereafter is what I had to do on my laptop.
# conda installation
v=3; f=Miniconda${v}-latest-MacOSX-x86_64.sh; cd $TMPDIR; { curl -LfOsS https://repo.anaconda.com/miniconda/$f ; cd -; } && bash $TMPDIR/$f -b && echo ". ~/miniconda${v}/etc/profile.d/conda.sh" >> ~/.bash_profile; . ~/miniconda${v}/bin/activateconda
# initialization
conda init
source ~/.bashrc
conda create --name py11 python==3.11
# activation
conda activate py11And hereafter the application;
import streamlit as st
import os
from pathlib import Path
from docling.backend.docling_parse_backend import DoclingParseDocumentBackend
from docling.datamodel.base_models import InputFormat
from docling.datamodel.pipeline_options import (
EasyOcrOptions,
OcrMacOptions,
PdfPipelineOptions,
RapidOcrOptions,
TesseractCliOcrOptions,
TesseractOcrOptions,
)
from docling.document_converter import DocumentConverter, PdfFormatOption
def main():
def main():
st.title("File Selection App")
# Initialize session state for storing the concatenated path
if 'full_file_path' not in st.session_state:
st.session_state.full_file_path = None
# Add description
st.write("Select a file from your computer to view its path information")
# File uploader with type filtering
allowed_types = ["pdf", "doc", "docx", "ppt", "pptx", "jpg", "jpeg"]
uploaded_file = st.file_uploader(
"Choose a file",
type=allowed_types,
help="Supported files: PDF, Word, PowerPoint, and JPEG"
)
# Display file details if a file is uploaded
if uploaded_file is not None:
# Get the current working directory
current_dir = os.getcwd()
# Create full path (this will be where Streamlit temporarily stores the file)
full_path = os.path.join(current_dir, uploaded_file.name)
# Store the concatenated path in session state
st.session_state.full_file_path = full_path
file_details = {
"Filename": uploaded_file.name,
"Full Path": full_path,
"File size": f"{uploaded_file.size / 1024:.2f} KB",
"File type": uploaded_file.type,
"Concatenated Path Variable": st.session_state.full_file_path
}
st.write("### File Details:")
for key, value in file_details.items():
st.write(f"**{key}:** {value}")
# Example of using the concatenated path in the app
st.write("### Using the Concatenated Path")
st.code(f"""
# You can access the full file path anywhere in your app using:
full_file_path = st.session_state.full_file_path
# Example usage:
if st.session_state.full_file_path:
# Do something with the path
print(f'Working with file: {st.session_state.full_file_path}')
""")
# Add a button to demonstrate using the path
if st.button("Print Path to Console"):
st.write(f"Path has been printed to console: {st.session_state.full_file_path}")
print(f"Full file path: {st.session_state.full_file_path}")
# Add a note about the path
st.info("Note: The full path shown is where Streamlit temporarily stores the uploaded file. "
"To get the original file path, you would need to use a different approach as browsers "
"don't provide the original file path for security reasons.")
######
##
input_doc = st.session_state.full_file_path
##
pipeline_options = PdfPipelineOptions()
pipeline_options.do_ocr = True
pipeline_options.do_table_structure = True
pipeline_options.table_structure_options.do_cell_matching = True
# Any of the OCR options can be used:EasyOcrOptions, TesseractOcrOptions, TesseractCliOcrOptions, OcrMacOptions(Mac only), RapidOcrOptions
# ocr_options = EasyOcrOptions(force_full_page_ocr=True)
# ocr_options = TesseractOcrOptions(force_full_page_ocr=True)
# ocr_options = OcrMacOptions(force_full_page_ocr=True)
# ocr_options = RapidOcrOptions(force_full_page_ocr=True)
ocr_options = TesseractCliOcrOptions(force_full_page_ocr=True)
pipeline_options.ocr_options = ocr_options
converter = DocumentConverter(
format_options={
InputFormat.PDF: PdfFormatOption(
pipeline_options=pipeline_options,
)
}
)
doc = converter.convert(input_doc).document
md = doc.export_to_markdown()
print(md)
if __name__ == "__main__":
main()In order to make the application distributable I set up the following “requirements.txt” (not the best type of requirements which could be written by a real Python developer 😅).
streamlit
pathlib
docling
logging
time
datetime
pandas
tensorflowTo test the app locally, the following simple command line does the job!
streamlit run app.pyAnd there goes the Dockerfile.
# Use an official Python runtime as a parent image
FROM python:3.11-slim
# Set environment variables
ENV PYTHONUNBUFFERED=1 \
PYTHONDONTWRITEBYTECODE=1 \
PIP_NO_CACHE_DIR=1
# Set the working directory in the container
WORKDIR /app
# Install system dependencies
RUN apt-get update && apt-get install -y \
build-essential \
curl \
software-properties-common \
git \
&& rm -rf /var/lib/apt/lists/*
# Copy only requirements first to leverage Docker cache
COPY requirements.txt .
# Install Python dependencies
RUN pip install --upgrade pip setuptools wheel \
&& pip install -r requirements.txt \
&& rm -rf /root/.cache/pip
# Copy the rest of the application
COPY . .
# Expose Streamlit port
EXPOSE 8501
# Add healthcheck
HEALTHCHECK CMD curl --fail http://localhost:8501/_stcore/health
# Set Streamlit configuration
ENV STREAMLIT_SERVER_PORT=8501
ENV STREAMLIT_SERVER_ADDRESS=0.0.0.0
# Run the application
CMD ["streamlit", "run", "app.py"]I use Podman to build and deploy the image, by aliasing Podman to Docker, all Docker commands could be used.
docker build -t app:latest .
#
docker run -p 8501:8501 app:latest
# or
docker run -d -p 8501:8501 app:latestConclusion
This article shows the ease of Docling implementation in order to make enterprise documents ready to be ingested easily for a RAG application for example.
Thanks for reading 🙏
Useful links
- Docling repository: https://github.com/DS4SD/docling
- Docling documentation and samples: https://ds4sd.github.io/docling/
- Docling PDF to Markdown sample: https://ds4sd.github.io/docling/examples/full_page_ocr/